
The best performance, with the lowest RMSD of 1.28, came from the SVM calculations. From the comparison between observed and theoretical, the researchers found a mostly poor performance from all five tools, with R 2 values ranging between 0.61 and 0.15. The second is made up of values obtained for the tryptic proteome generated from the cellular fraction of Drosophila Kc167 cells.įor the theoretical values generated for proteins, the team first grouped the results into those with variable p Is and those with only one unique p I. From this analysis, they found that most proteins do not possess a unique pI. used two databases for reference the first, the PIP-DB ( protein isoelectric point database) contains a comprehensive record of protein p I data. The researchers then calculated p I values using each of the tools under investigation before comparing the theoretical results obtained against those publicly available. In this way, the benchmarking process would allow for direct comparisons through reference to correlation and root-mean-square deviation (RMSD) evaluation. note that in order to avoid bias in reporting, they did not optimize the methods used for evaluation for any of the tools under investigation.įirst, the team constructed an R-package, a collection of programs, functions and data written in statistical programming language R, as a framework for reproducible analysis within which to examine performance of the various algorithms. Branca: calculation according to correction factors for position, influence of neighboring groups, and statistical corrections for presence and nature of side chain groupsĪudain et al.Support Vector Machine (SVM): calculation based on amino acid sequence and Amino Acid Index database ( AAindex) data.Bjellqvist: calculated according to pKa and amino acid position.Cofactor: calculated with correction factors according to amino acid position and adjacent charged residues.Iterative: calculated from amino acid sequence.The researchers chose the following tools to undergo benchmarking:


1 The researchers benchmarked algorithm performance, comparing results obtained against public data sets to show how well these predictive tools performed. (2015) compared and contrasted five tools available to researchers for determining p I on the basis of amino acid sequence. Although these predictive methods exist, their performance can be variable and may skew ensuing results.Īudain et al. Various methods for predicting p I in denatured proteins exist, and most base this calculation on amino acid sequence with reference to p K a values recorded for ionizable constituents. p I depends on a number of factors, including amino acid sequence, post-translational modifications (PTMs) and presence of side chain-all of which can alter surface charge and behavior depending on the pH of the environment.
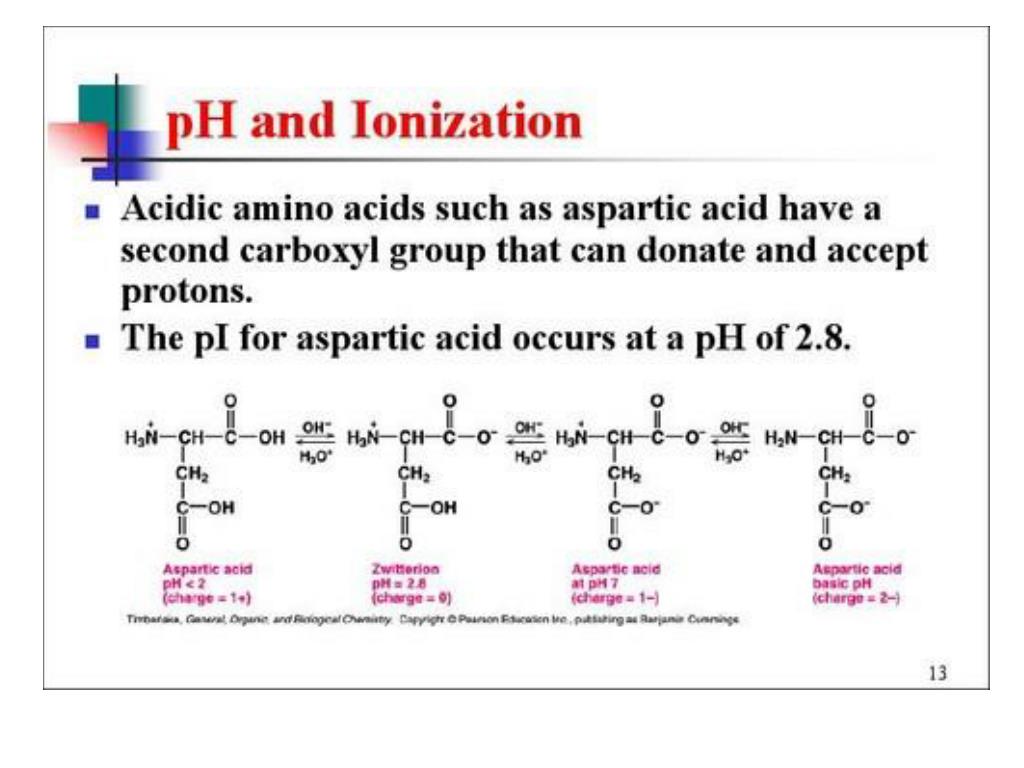
This factor governs electrophoretic mobility in proteins and also plays a role in identifying peptides from mass spectral proteomics data. Or from a ProtParam.The isoelectric point, or p I,represents a point of balance for a molecule, where the external surface charge is a net zero. The methods of this class can either be accessed from the class itself Occurences as integers, e.g.ĭefault: None.

:aa_content: A dictionary with amino acid letters as keys and it’s Parameters :protein_sequence: A ``Bio.Seq`` or string object containing a protein IsoelectricPoint ( protein_sequence, aa_content = None ) ¶Ī class for calculating the IEP or charge at given pH of a protein. I designed the algorithm according to a note by David L. Reference points for comparisons of two - dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences.


 0 kommentar(er)
0 kommentar(er)
